My long-term research interest lies with understanding how and why biological macromolecules evolve at characteristic -- and hugely different -- rates. As a result of some great work in the field of molecular evolution in the latter half of the twentieth century, there is the perception that we understand fairly well on a molecular level how the forces of mutation and natural selection combine to cause the sequences of genes and proteins to change over time. People have even created detailed mathematical models that simulate patterns of sequence evolution over millions of years. And in general they work pretty well.
As we understand the process, the rate of molecular evolution should be dependent on (a) the rate at which mutations occur in an organism's genes, and (b) the rate at which these mutations become fixed in a population of organisms, either because the mutations are advantageous or because they are not disadvantageous. In the simplest model, the forces of mutation and selection are the same for all organisms across the tree of life and across evolutionary time.
This may be a reasonable starting point, but it's rather naive. It's easy to imagine that different types of organisms might experience different mutational environments, and any student of biochemistry can tell you that different proteins have different constraints on their structure due to their different functions. Nevertheless, it seems reasonable to postulate that a given gene or protein should experience similar selective pressure to conserve its structure and function across most organisms under the evolutionary equivalent of the law
Starting in the 1990s and continuing at an ever-increasing pace, we've seen a huge increase in the amount of data on sequences of genes from diverse organisms, providing a great way to test our understanding of the processes of molecular evolution. And as we look through this mountain of data, it becomes increasingly clear that the simple view of evolutionary rates simply does not apply to many genes in many organisms. That is to say, something very exciting is going on -- there are forces at work that we don't understand!
So how does one get at these mysterious forces? A geneticist seeks to understand biological function by examining the factors underlying a mutant phenotype. In the same vein, my approach to understanding the regulation of evolutionary rates is to dissect the factors which underlie natural occurrences of radically altered gene evolution. These are my "molecular evolutionary mutants", or, if you will, evidence of Mother Nature's dabbling in Experimental Evolutionary Biochemistry. My students and I are currently working on the elucidation of one such "experiment", involving the genes that encode the RNA polymerase in the chloroplasts of plants in the genus Pelargonium.
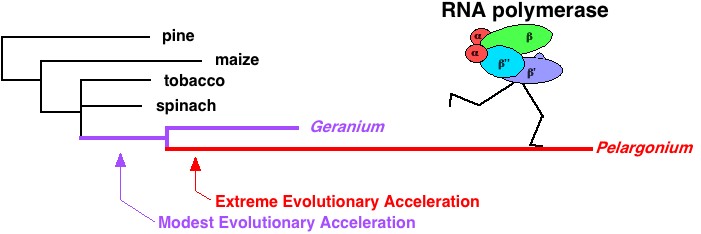
A dramatic acceleration -- somewhere between ten- and one hundred-fold -- in evolutionary rate distinguishes the genes encoding the chloroplast RNA polymerase in the relatives of the common garden geranium, Pelargonium hortorum, from those of other flowering plants. My initial work on this project was done in Jeff Palmer's lab at Indiana University, building on the work of John Logsdon (now at the University of Iowa) and Pat Calie (now at Eastern Kentucky U.). The most exciting aspect of these studies was evidence that at least one of the rpo genes, encoding one of the catalytic subunits of the RNA polymerase, is under strong positive selection for amino acid sequence change. That is to say that natural selection, for as yet unknown reasons, seems to actively favor the reproductive success of Pelargonium plants whose RNA polymerase subunits are evolving very rapidly. Incredibly, we have evidence for a more marked level of selection than the "textbook" case of positive Darwinian selection, involving the Complementarity Determining Regions of mammalian immunoglobulins -- where it is clearly in an organism's favor to foster sequence diversity so as to recognize diverse antigens!
At Denison, I've continued to flesh out the rate of nucleotide sequence evolution (with the help of Punya Navaratnarajah (DU 2007) and Kristin French (DU 2009)), and also embarked on a detailed biochemical and molecular biological investigation of the consequences of the extremely rapid evolution on the function of the Pelargonium chloroplast RNA polymerase. Over time, this investigation should give us insights into the relationships between protein sequence and function, and hopefully will lead us toward an understanding of what evolutionary agent has driven the Pelargonium RNA polymerases to evolve so rapidly.
Much of our work is focused on developing the systems we will use to test the activity of the polymerases and then test our hypotheses about protein sequence/function relationships. With the help of Todd Wine (DU class of 1999), Heidi Duff (DU 2000), Brittany Gagne (DU 2002), and Ashley Galant (DU 2006), I developed a novel non-radioisotopic assay for RNA polymerase activity. Having found a robust and sensitive way to measure the functional activity of our enzyme, our primary focus has shifted to purification of the active polymerase. Greg Mecoli (DU 2006), Ashley Galant (DU 2006) and Alan Roach (DU 2004) took initial steps towards the purification of the RNA polymerase(s) from Pelargonium leaves. Purified Pelargonium polymerase should allow us (a) to confirm that the unusual gene sequences are mirrored in the sequences of the subunits from the transcriptionally competent enzyme, and (b) see if its enzymatic properties match those of RNA polymerases from other plants whose polymerase-encoding genes are evolving at a more conventional rate.
To complement and extend our work with the enzyme isolated from Pelargonium tissue, we have also been developing a purely ex vivo system for probing the effects of the rapid polymerase evolution. Chrissy Gorman (DU 2001) successfully replicated published work from investigators of the E. coli RNA polymerase in preparation for reconstituting "engineered" Pelargonium polymerases in vitro, and Nick Ostrout (DU 2003) and Ashley Galant have advanced the Pelargonium reconstitution effort by cloning several of the RNA polymerase-encoding genes from Pelargonium hortorum into expression cassettes.
Both the "natural" and reconstituted polymerases should provide insight into the characteristics of transcription under the control of the rapidly-evolving Pelargonium RNA polymerases. The third active route of investigation in the lab aims to provide complementary information: what is the pattern of gene expression by Pelargonium RNA polymerases in living plant tissue? To get some preliminary answers to that question, Ezinnaya Ubagharaji (DU 2009), Bethany Wood (DU 2006) and Ann Staubach (DU 2004) have taken steps toward development of an array of probes for assaying chloroplast gene expression patterns under conditions where the rapidly-evolving Pelargonium RNA polymerase is, and is not, expressed. By comparing in vivo and in vitro expression patterns of Pelargonium and control polymerases, and by examining chloroplast gene promoter sequences in Pelargonium and other angiosperms, we hope to learn more about the functional consequences of the accelerated evolution.
Finally, Marcus Breese (DU 2000) worked to develop a computer program that would help us to generate three-dimensional models of protein structures based on natural sequence variations. In collaboration with Pat Calie's and Debra Bautista's groups at Eastern Kentucky, this project is continuing with new tools, as we model possible chloroplast RNA polymerase structure based on the structure of the homologous RNA polymerase from Thermus aquaticus. These models will guide the selection of mutagenesis experiments to probe Pelargonium RNA polymerase structure/function/evolution relationships in our in vitro system.
If you are considering conducting research with me, I encourage you to come by and talk with me about your interests and options! You may also wish to look over my expectations for student research projects under my supervision.